Welcome to Git@WUR
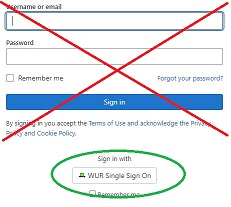
Login
External users: use the username/password form.
WUR users: use the button 'WUR Single Sign On'.
Two-factor authentication (2FA)
Please enable two-factor authentication if not already enabled.
For general information go to ‘Manage your code with Git@WUR’.
For technical information go to ‘Help files Git@WUR’.